"encouraging psycholinguists to use linear mixed-effects models [is] like giving shotguns to toddlers” (Prominent psycholinguist cited by Barr et al. 2013)
Benefits of LMMs¶
- Extending linear models to within design
- Data pooling, learning about one participants informs you about the other participant
- No pre-averaging
- Accounting for imbalance in sampling
Costs of LMMs¶
- More decisions have to be made
- Hard to understand as we have multiple levels
- Implement and computation power
Organization of the second module¶
- Maximum likelihood estimation
- What are hierarchies in the data
- Formalism behind LMM
- Illustrating the concept of random effects
- Adding predictor
- Full LMM
We are going to flip coins to illustrate the concept
What is the probability of $x$ number of heads given $n$ coin flips
$$ P(x|n,p) = \frac{n!}{x!(n-x)!} \cdot p^x(1 - p)^{n-x} $$If I come up with 28 heads for 40 flips:
With assumpation that the coin is unbiased, i.e. $p=0.5$ :
\begin{equation} P(28|40,.5) = \frac{40!}{28!(40-28)!} \cdot .5^{28}(1 - .5)^{40-28} \approx .002 \end{equation}with $p=0.7$ \begin{equation} P(28|40,.7) = \frac{40!}{28!(40-28)!} \cdot .70^{28}(1 - .7)^{40-28} \approx .137 \end{equation}
Given these results, what is the most likely ? unbiased coin or coin biased towards heads ?
Now moving to continuous DVs same logic applies except that we are not looking at probability mass functions but probability density functions, e.g. density of a normal distribution :
$$ f(x) = \frac{1}{\sigma\sqrt{2\pi}} \exp\left( -\frac{1}{2}\left(\frac{x-\mu}{\sigma}\right)^{\!2}\,\right) $$
We are looking for the parameter values (e.g. $\mu$) that maximises the likelihood of the data. E.g. :
dnorm(100,100,15)#probability density function for an IQ of 100 given IQ~N(100,15)
vs.
dnorm(100,70,15)#Same but given IQ~N(70,15)
This, obviously in a more complex way, is the method under the hood of parameter estimation in the LMMs we are going to study : we optimize (maximize) the likelihood function of our models.
What are hierarchies in the data ?¶
Distributions of parameters of a distribution
#code was directly inspired by S. Kurz, see https://solomonkurz.netlify.app/post/make-model-diagrams-kruschke-style/
(p1_1 + p1_2 + p2_1 + p2_2 + p3 + p4 + p5) +
plot_layout(design = layout) &
ylim(0, 1)
theme_set(theme_grey())#Reset ggplot theme,
Let's create a data set with hierarchies¶
Imagine a WM task where we sampled $x$ WM scores by participant $\times$ condition (within design and repeated measure)
We want to create participants which present an inter-individual variability, therefore we define a distribution on the parameter $\mu$ so that each individual receives a unique $\mu$ parameter
The distribution we will sample from is given below :
###### Distributions mu parameter
x = seq(2, 10, length=1000)
plot(x, dnorm(x, mean=6, sd=1), type="l", ylab="density", xlab="mu value")
Now we create a for loop were we generate up to 100 trials for 15 participants, each run draws random values for the $\mu$ parameter and a random number of trial (e.g. a priori rejection criterion, fatigue,...)
mean_wm = 6
sd_wm = 1
set.seed(234)
n_participants = 15
data = data.frame()
ntrial_v = NULL
mu_v = NULL
for(i in 1:n_participants){
ntrials = runif(1, 5, 100)# Ntrials from 5 to 100
subdata = data.frame(trial=1:ntrials)#trial number vector
mu = rnorm(1, mean=mean_wm, sd=sd_wm)#random sample from the normal illustrated above
subdata$wm = rnorm(ntrials, mu, sd=sd_wm)#generating data based on the drawn mu
subdata$participant = as.character(i) #recording participant
data = rbind(data, subdata)#pasting the new subject to the df
ntrial_v[i] <- ntrials#SAving the number of trials per participant
}
tail(data)
| trial | wm | participant | |
|---|---|---|---|
| <int> | <dbl> | <chr> | |
| 814 | 30 | 7.437569 | 15 |
| 815 | 31 | 7.642065 | 15 |
| 816 | 32 | 7.408912 | 15 |
| 817 | 33 | 9.127590 | 15 |
| 818 | 34 | 7.106579 | 15 |
| 819 | 35 | 7.074170 | 15 |
We can look at the n of trials we sampled
hist(ntrial_v,breaks=30)
We can take a look at the distribution of WM for all the participants
hist(data$wm, breaks=100, xlim=c(3,9))
or for each participant
require(lattice)#needed for histogram plot
options(repr.plot.width=10, repr.plot.height=10)
histogram( ~ wm | participant ,data = data, breaks=20)
options(repr.plot.width=8, repr.plot.height=5, repr.plot.res = 200)
Loading required package: lattice
Adding experimental levels¶
We will now add an experimental condition, e.g. imagine we have a perfect longitudinal study on age and working memory, we tested each individual again at different ages
library(MASS)#we will sample from a multivariate normal distribution
data = data.frame()
varcovmat = matrix(c(1, 0, 0, 1), nrow=2)#See later for var-cov matrix
data = data.frame()
for(i in 1:n_participants){
re = mvrnorm(n=1, mu=c(0, 0), Sigma=varcovmat)#Sampling from the mvn, rescaling according to DV distribution in next lines
mu_i = re[1] * sd_wm + mean_wm#participant keeps the same mu during all the years
b_age = re[2] *(sd_wm*2) - (sd_wm*4) ##Each participant get a single slope for age, the effect size is displayed as a unit of sd_wm
age_tested = runif(runif(1, 5,15), 1, 99)#retesting participant at different random [1,99] ages [5,15] times
for (age in age_tested){# A for loop in a for loop is especially dirty but more transparent
age = age/100#We recode age for scale purpose
ntrials = runif(1, 5, 100)
subdata = data.frame(trial=1:ntrials)
mu = mu_i+b_age*age#inducing age diff alpha + beta * IV
subdata$wm = rnorm(ntrials, mu, sd=sd_wm)
subdata$participant = as.character(i)
subdata$age = age #storing age
data = rbind(data, subdata)
}
}
Attaching package: ‘MASS’
The following object is masked from ‘package:patchwork’:
area
The following object is masked from ‘package:dplyr’:
select
For a starter we are going to pre-average the data
means_bysub = aggregate(data$wm, FUN=mean,
by=list(age=data$age, participant = data$participant))
library(ggplot2)
ggplot() +
geom_point(data=means_bysub, aes(x=age, y=x, group = participant, colour = participant)) +
scale_colour_discrete('participant')
A simple LM :
m0 <- lm(x ~ age, data = means_bysub)#Syntax discussed later
means_bysub$random.intercept.preds <- predict(m0)
options(repr.plot.width=7, repr.plot.height=5, repr.plot.res = 300)
ggplot() +
geom_point(data=means_bysub, aes(x=age, y=x, group = participant, colour = participant)) +
geom_line(data=means_bysub, aes(x=age, y=random.intercept.preds)) +
labs(x="age", y="WM") +
scale_colour_discrete('participant')
To respect assumptions of LM we need i.i.d. data :
- We could fit on lm() per participant -> unefficient and how do we perform the second step about population effect ?
- We can fit linear mixed models
Formalism behind LMM¶
Mixed models are said mixed because they include fixed effects and random effects (confusing terminology: multilevel, hierarchical, random,...)
- Fixed = our effects of interest (e.g. age)
- Random = an effect (e.g. inter-individual variations) not in the scope of our research question
LMM are extensions of LM :
$y_{ji} \sim \mathcal{N}(\mu_j, \sigma)$
But this time :
$\mu_j = \alpha_j + \beta_j IV$
$\alpha_j \sim \mathcal{N}(\mu_\alpha, \sigma_\alpha)$
$\beta_j \sim \mathcal{N}(\mu_\beta, \sigma_\beta)$
As in module 1, adding predictors just increase the number of $\beta_j$
$\alpha_j \sim \mathcal{N}(\mu_\alpha, \sigma_\alpha)$
$\beta_j \sim \mathcal{N}(\mu_\beta, \sigma_\beta)$
As in module 1, adding predictors just increase the number of $\beta_j$
mu_j = rnorm(1, mean=mean_wm, sd=sd_wm)#Random intercept a_j
b_age_j = rnorm(1, -4, 2)#Random slope
Illustrating the concept of random effects¶
We are now going to use (one of) the R package allowing to fit mixed models : lme4
#install.packages(lme4)
library(lme4)
Loading required package: Matrix
Attaching package: ‘Matrix’
The following objects are masked from ‘package:tidyr’:
expand, pack, unpack
Next we illustrate the notion of random intercept vs random slopes vs full random effects (R code used was largely inspired by : http://mfviz.com/hierarchical-models/ )
A model with random effects on the intercept ($\alpha_j$)¶
m1 <- lmer(x ~ age + (1|participant), data = means_bysub)#Syntax
means_bysub$random.intercept.preds <- predict(m1)
library(ggplot2)
options(repr.plot.width=7, repr.plot.height=5, repr.plot.res = 300)
ggplot() +
geom_point(data=means_bysub, aes(x=age, y=x, group = participant, colour = participant)) +
geom_line(data=means_bysub, aes(x=age, y=random.intercept.preds, group = participant, colour = participant)) +
labs(x="age", y="WM") +
scale_colour_discrete('participant')
A model with random effects on the slope ($\beta_j$)¶
m2 <- lmer(x ~ age + (0+age|participant), data = means_bysub)
means_bysub$random.intercept.preds <- predict(m2)
options(repr.plot.width=7, repr.plot.height=5, repr.plot.res = 300)
ggplot() +
geom_point(data=means_bysub, aes(x=age, y=x, group = participant, colour = participant)) +
geom_line(data=means_bysub, aes(x=age, y=random.intercept.preds, group = participant, colour = participant)) +
labs(x="age", y="WM") +
scale_colour_discrete('participant')
A model with random effects on the intercept and on the slope ($\alpha_j$ and $\beta_j$)¶
m3 <- lmer(x ~ age + (age||participant), data = means_bysub)#1+ is implicit, 0+ explicitely removes intercept
#In this model I specified || which removes the estimation of the correlation between intercept and slope, more later
means_bysub$random.intercept.preds <- predict(m3)
options(repr.plot.width=7, repr.plot.height=5, repr.plot.res = 300)
ggplot() +
geom_point(data=means_bysub, aes(x=age, y=x, group = participant, colour = participant)) +
geom_line(data=means_bysub, aes(x=age, y=random.intercept.preds, group = participant, colour = participant)) +
labs(x="age", y="WM") +
scale_colour_discrete('participant')
Illustrating shrinkage¶
We will compare parameter estimate from one lm() / participant vs the partial pooling operated by LMMs
i_slope_values = NULL
i_intercept_values = NULL
for(i in 1:length(unique(means_bysub$participant))){
m0_ = lm(x ~ age, data = means_bysub[means_bysub$participant == i,])
i_slope_values[i] = m0_$coefficients["age"]
i_intercept_values[i] = m0_$coefficients["(Intercept)"]
}
h_slope_values = coef(m3)$participant$age
h_intercept_values = coef(m3)$participant$'(Intercept)'
plot(i_intercept_values, i_slope_values, xlab="Intercept", ylab="Slope")#parameter of individual LM
points(h_intercept_values, h_slope_values, col="red")#FE parameters of LMM
abline(v=6)#True intercept
abline(h=-4)#True slope
To go further on the notion of pooling (one LM), vs. partial pooling (LMM) vs. no-pooling (one LM / participant) see the blog post by T.Mahr
Inspecting and comparing the outputs¶
summary(m0)
Call:
lm(formula = x ~ age, data = means_bysub)
Residuals:
Min 1Q Median 3Q Max
-3.3510 -1.2478 -0.0826 1.6419 2.6184
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 5.8117 0.2766 21.012 < 2e-16 ***
age -2.9681 0.4701 -6.313 3.72e-09 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.576 on 134 degrees of freedom
Multiple R-squared: 0.2292, Adjusted R-squared: 0.2235
F-statistic: 39.86 on 1 and 134 DF, p-value: 3.72e-09
summary(m1)
Linear mixed model fit by REML ['lmerMod']
Formula: x ~ age + (1 | participant)
Data: means_bysub
REML criterion at convergence: 261.1
Scaled residuals:
Min 1Q Median 3Q Max
-2.46308 -0.47152 -0.00825 0.53307 2.50096
Random effects:
Groups Name Variance Std.Dev.
participant (Intercept) 2.0696 1.4386
Residual 0.2491 0.4991
Number of obs: 136, groups: participant, 15
Fixed effects:
Estimate Std. Error t value
(Intercept) 5.6974 0.3825 14.90
age -2.9445 0.1553 -18.96
Correlation of Fixed Effects:
(Intr)
age -0.207
summary(m2)
Linear mixed model fit by REML ['lmerMod']
Formula: x ~ age + (0 + age | participant)
Data: means_bysub
REML criterion at convergence: 306.2
Scaled residuals:
Min 1Q Median 3Q Max
-2.48144 -0.50664 -0.07217 0.53755 2.44197
Random effects:
Groups Name Variance Std.Dev.
participant age 6.9532 2.6369
Residual 0.3591 0.5993
Number of obs: 136, groups: participant, 15
Fixed effects:
Estimate Std. Error t value
(Intercept) 5.9566 0.1078 55.265
age -3.5092 0.7085 -4.953
Correlation of Fixed Effects:
(Intr)
age -0.237
summary(m3)
Linear mixed model fit by REML ['lmerMod']
Formula: x ~ age + ((1 | participant) + (0 + age | participant))
Data: means_bysub
REML criterion at convergence: 52.5
Scaled residuals:
Min 1Q Median 3Q Max
-4.5813 -0.5184 0.1037 0.5889 1.7286
Random effects:
Groups Name Variance Std.Dev.
participant (Intercept) 1.1721 1.0826
participant.1 age 3.4357 1.8536
Residual 0.0287 0.1694
Number of obs: 136, groups: participant, 15
Fixed effects:
Estimate Std. Error t value
(Intercept) 5.8333 0.2822 20.672
age -3.3565 0.4831 -6.947
Correlation of Fixed Effects:
(Intr)
age -0.017
"Significance" testing¶
When focusing on significance testing (but also when focus is on estimation) you should carefully think about the contrast you used for your factors (cf. module 1).
e.g. :
- how could my intercept make sense in a significance testing framework ?
- Am I interested in the difference between two levels (sum-coding, [-0.5, 0.5]) ? Or how moving from one standard condition to the other changes the intercept (treatment coding, [0,1])
How do I assess significance of my predictors ?
- p values but not that obvious to compute because of the estimation of degrees of freedom
- Rule on t values (e.g. > 1.96) also implies an assumption on DoF
- 0 excluded from CI, CI might not have the definition you think they have (but more on that in Module 3)
#install.packages('lmerTest')
library('lmerTest')#provides estimation of DoF and Pvalues
Attaching package: ‘lmerTest’
The following object is masked from ‘package:lme4’:
lmer
The following object is masked from ‘package:stats’:
step
m1 <- lmer(x ~ age + (1|participant), data = means_bysub)
m2 <- lmer(x ~ age + (0+age|participant), data = means_bysub)
m3 <- lmer(x ~ age + (age||participant), data = means_bysub)
summary(m1)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: x ~ age + (1 | participant)
Data: means_bysub
REML criterion at convergence: 261.1
Scaled residuals:
Min 1Q Median 3Q Max
-2.46308 -0.47152 -0.00825 0.53307 2.50096
Random effects:
Groups Name Variance Std.Dev.
participant (Intercept) 2.0696 1.4386
Residual 0.2491 0.4991
Number of obs: 136, groups: participant, 15
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 5.6974 0.3825 15.3306 14.90 1.57e-10 ***
age -2.9445 0.1553 120.4660 -18.96 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr)
age -0.207
summary(m2)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: x ~ age + (0 + age | participant)
Data: means_bysub
REML criterion at convergence: 306.2
Scaled residuals:
Min 1Q Median 3Q Max
-2.48144 -0.50664 -0.07217 0.53755 2.44197
Random effects:
Groups Name Variance Std.Dev.
participant age 6.9532 2.6369
Residual 0.3591 0.5993
Number of obs: 136, groups: participant, 15
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 5.9566 0.1078 120.0983 55.265 < 2e-16 ***
age -3.5092 0.7085 15.4526 -4.953 0.000159 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr)
age -0.237
summary(m3)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: x ~ age + (age || participant)
Data: means_bysub
REML criterion at convergence: 52.5
Scaled residuals:
Min 1Q Median 3Q Max
-4.5813 -0.5184 0.1037 0.5889 1.7286
Random effects:
Groups Name Variance Std.Dev.
participant (Intercept) 1.1721 1.0826
participant.1 age 3.4357 1.8536
Residual 0.0287 0.1694
Number of obs: 136, groups: participant, 15
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 5.8333 0.2822 14.1448 20.672 5.71e-12 ***
age -3.3565 0.4831 13.9238 -6.947 6.99e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr)
age -0.017
Full random effect structure does not necessarly imply higher power but it does (as good as possible) guard against Type I errors, see Barr et al., 2013
variance/covariance matrix¶
Note that the last model, including two random effects, did not estimate a correlation among those random effects. However usually this could be true (e.g. participants with higher scores/intercept might be less subject to age decline/slope) and we would like to estimate it.
This is best represented by a variance-covariance matrix :
$$\begin{pmatrix} \text{Var(Intercept)} & \text{Cov(Intercept,Slope)} \\\ \text{Cov(Intercept,Slope)} & \text{Var(Slope)} \end{pmatrix}$$Let's recreate the data with a defined var-covariance matrix : $$\begin{pmatrix} 1 & 0.66 \\ 0.66 & 2 \end{pmatrix}$$
library(MASS)#we will sample from a multivariate normal distribution
varcovmat = matrix(c(1, .66, .66, 2), nrow=2)#here covar = 0.66 = correlation of .33
matrix_mvn = mvrnorm(10000, mu=c(6, -4), Sigma=varcovmat)
plot(matrix_mvn[,1],matrix_mvn[,2], xlab = "Intercept", ylab = "Slope")
data = data.frame()
varcovmat = matrix(c(1, .66, .66, 2), nrow=2)#Same covar matrix
data = data.frame()
for(i in 1:n_participants){
re = mvrnorm(n=1, mu=c(6, -1), Sigma=varcovmat)#Sampling from the mvn, rescaling according to DV distribution in next lines
mu_i = re[1]
b_age = re[2]
age_tested = runif(runif(1, 5,15), 1, 99)#retesting participant at different random [1,99] ages [5,15] times
for (age in age_tested){# A for loop in a for loop is especially dirty but more transparent
age = age/100#We recode age for scale purpose
ntrials = runif(1, 5, 100)
subdata = data.frame(trial=1:ntrials)
mu = mu_i+b_age*age#inducing age diff alpha + beta * IV
subdata$wm = rnorm(ntrials, mu, sd=sd_wm)
subdata$participant = as.character(i)
subdata$age = age #storing age
data = rbind(data, subdata)
}
}
m3_cov <- lmer(x ~ age + (age|participant), data = means_bysub)#One | only indicates we estimate the covariance between RE
summary(m3_cov)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: x ~ age + (age | participant)
Data: means_bysub
REML criterion at convergence: 52.3
Scaled residuals:
Min 1Q Median 3Q Max
-4.5775 -0.5253 0.0993 0.5893 1.7260
Random effects:
Groups Name Variance Std.Dev. Corr
participant (Intercept) 1.16819 1.0808
age 3.42230 1.8499 0.13
Residual 0.02871 0.1694
Number of obs: 136, groups: participant, 15
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 5.8338 0.2817 14.1429 20.708 5.59e-12 ***
age -3.3568 0.4822 13.9188 -6.961 6.85e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr)
age 0.110
In our model the estimation made of the covariance matrix is = $$\begin{pmatrix} 1.17 & 1.32 \\ 1.32 & 3.42 \end{pmatrix}$$
Note that the random effect variances and covariances are difficult to estimate when we have small sample sizes (as always by the way, e.g. for correlation see blog post by G. Rousselet)
Adding another predictor¶
#First we define our covariance matrix unitless, we later rescale to the values of the DV
varcovmat = matrix(c(1, .33, .33, .33, .33, 1, .33, .33, .33, .33,1,.33, .33, .33, .33,1), nrow=4)#everything is correlated at .33
data = data.frame()
for(i in 1:n_participants){
re = mvrnorm(n=1, mu=c(0, 0, 0, 0), Sigma=varcovmat)
mu_i = re[1] * sd_wm + mean_wm #Rescaling so that mean = 6 and sd = 1
b_age = re[2] *(sd_wm*2) - (sd_wm*4) #b_age_j + rescaling
b_task = re[3] * sd_wm + -(sd_wm*2)#b_task_j + rescaling
b_age_task = re[4] * (sd_wm/2) - (sd_wm*2)#b_agetask_j + rescaling
age_tested = runif(runif(1, 5,15), 1, 99)
for (age in age_tested){# for loop in for loops is especially dirty but more transparent
age = age/100#We recode age for scale purpose
for (task in c(0,1)){
ntrials = runif(1, 50, 150)
subdata = data.frame(trial=1:ntrials)
mu = mu_i+b_age*age+b_task*task+b_age_task*age*task
subdata$wm = rnorm(ntrials, mu, sd=sd_wm)
subdata$participant = as.character(i)
subdata$age = age
subdata$task = task
data = rbind(data, subdata)
}
}
}
means_ = aggregate(data$wm, FUN=mean,
by=list(age=data$age, task=data$task, participant = data$participant))#pre-averaging at first
m4 <- lmer(x ~ age * task + (1+age*task|participant), data = means_)
Warning message in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, : “Model failed to converge with max|grad| = 0.0028704 (tol = 0.002, component 1)”
We have a convergence error, max likelihood optimizer is having some trouble converging (when you get convergence problems, see this page).
In our case we can simply change the optimization criterion
library(optimx)
m4 <- lmer(x ~ age * task + (1+age*task|participant), data = means_,
control = lmerControl(optimizer = "optimx", calc.derivs = FALSE,
optCtrl = list(method = "nlminb", starttests = FALSE, kkt = FALSE)))
But warnings can be informative about the structure of the LMM we are trying to fit, e.g. we could have more parameters than observations
#m4_wrong <- lmer(x ~ age * task + (1+age*task|participant), data = means_[means_$age < .1,])
Problem is also the number of units in your random effects. The higher the number the better will be the estimation of variance (and co-variance !) of random effects
summary(m4)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: x ~ age * task + (1 + age * task | participant)
Data: means_
Control:
lmerControl(optimizer = "optimx", calc.derivs = FALSE, optCtrl = list(method = "nlminb",
starttests = FALSE, kkt = FALSE))
REML criterion at convergence: -83.7
Scaled residuals:
Min 1Q Median 3Q Max
-2.62389 -0.58975 -0.00013 0.56033 3.01961
Random effects:
Groups Name Variance Std.Dev. Corr
participant (Intercept) 1.147251 1.07110
age 3.118870 1.76603 0.48
task 0.752606 0.86753 0.50 0.44
age:task 0.261845 0.51171 0.56 0.23 0.27
Residual 0.009878 0.09939
Number of obs: 216, groups: participant, 15
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 6.0378 0.2774 14.0103 21.768 3.35e-12 ***
age -3.6195 0.4574 13.9459 -7.914 1.59e-06 ***
task -1.8672 0.2260 13.9988 -8.263 9.38e-07 ***
age:task -2.0807 0.1413 14.6248 -14.725 3.58e-10 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) age task
age 0.470
task 0.486 0.438
age:task 0.541 0.196 0.209
Plotting the multiple predictor model¶
#install.packages("sjPlot")
library("sjPlot")
library(sjlabelled)
library(sjmisc)
library(ggplot2)
Attaching package: ‘sjlabelled’
The following object is masked from ‘package:forcats’:
as_factor
The following object is masked from ‘package:dplyr’:
as_label
Attaching package: ‘sjmisc’
The following object is masked from ‘package:purrr’:
is_empty
The following object is masked from ‘package:tidyr’:
replace_na
The following object is masked from ‘package:tibble’:
add_case
plot_model(m4, type = "pred", terms = c("age", "task"), show.data = TRUE)
#install.packages('glmmTMB')
plot_model(m4, type = "est")
#install.packages('glmmTMB')
plot_model(m4, type = "re")
Now checking models assumption¶
library(performance)
options(repr.plot.width=8, repr.plot.height=10, repr.plot.res = 100)#hidden code for display size
check_model(m4)
options(repr.plot.width=8, repr.plot.height=5, repr.plot.res = 200)#hidden code for display size
Loading required namespace: qqplotr For confidence bands, please install `qqplotr`.
Fitting a LMM at the trial level¶
Here we build a linear mixed model with all fixed effects (simple and interaction) and all random effects (intercept and all effects) at the trial level (without pre-averaging)
-> We use the model with the maximum of information where averaging is part of the model
full_trial_fullFE <- lmer(wm ~ age * task + (age*task|participant), data = data,
control = lmerControl(optimizer = "optimx", calc.derivs = FALSE,
optCtrl = list(method = "nlminb", starttests = FALSE, kkt = FALSE)))
summary(full_trial_fullFE)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: wm ~ age * task + (age * task | participant)
Data: data
Control:
lmerControl(optimizer = "optimx", calc.derivs = FALSE, optCtrl = list(method = "nlminb",
starttests = FALSE, kkt = FALSE))
REML criterion at convergence: 60887.6
Scaled residuals:
Min 1Q Median 3Q Max
-3.6648 -0.6854 0.0110 0.6718 3.5132
Random effects:
Groups Name Variance Std.Dev. Corr
participant (Intercept) 1.1317 1.0638
age 3.0951 1.7593 0.48
task 0.7551 0.8690 0.51 0.44
age:task 0.2507 0.5007 0.58 0.24 0.30
Residual 1.0123 1.0061
Number of obs: 21260, groups: participant, 15
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 6.0423 0.2756 14.0087 21.923 3.05e-12 ***
age -3.6329 0.4558 13.9188 -7.970 1.49e-06 ***
task -1.8768 0.2266 14.0056 -8.284 9.08e-07 ***
age:task -2.0651 0.1394 14.4719 -14.810 3.83e-10 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) age task
age 0.475
task 0.493 0.441
age:task 0.559 0.197 0.227
summary(m4)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: x ~ age * task + (1 + age * task | participant)
Data: means_
Control:
lmerControl(optimizer = "optimx", calc.derivs = FALSE, optCtrl = list(method = "nlminb",
starttests = FALSE, kkt = FALSE))
REML criterion at convergence: -83.7
Scaled residuals:
Min 1Q Median 3Q Max
-2.62389 -0.58975 -0.00013 0.56033 3.01961
Random effects:
Groups Name Variance Std.Dev. Corr
participant (Intercept) 1.147251 1.07110
age 3.118870 1.76603 0.48
task 0.752606 0.86753 0.50 0.44
age:task 0.261845 0.51171 0.56 0.23 0.27
Residual 0.009878 0.09939
Number of obs: 216, groups: participant, 15
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 6.0378 0.2774 14.0103 21.768 3.35e-12 ***
age -3.6195 0.4574 13.9459 -7.914 1.59e-06 ***
task -1.8672 0.2260 13.9988 -8.263 9.38e-07 ***
age:task -2.0807 0.1413 14.6248 -14.725 3.58e-10 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) age task
age 0.470
task 0.486 0.438
age:task 0.541 0.196 0.209
plot_model(full_trial_fullFE, type = "pred", terms = c("age", "task"), show.data = FALSE)
plot_model(full_trial_fullFE)
plot_model(full_trial_fullFE, type="re")
And the assumption of our model ??¶
#check_model(full_trial_fullFE)# eats too much RAM and therefore too slow so I saved it in advance
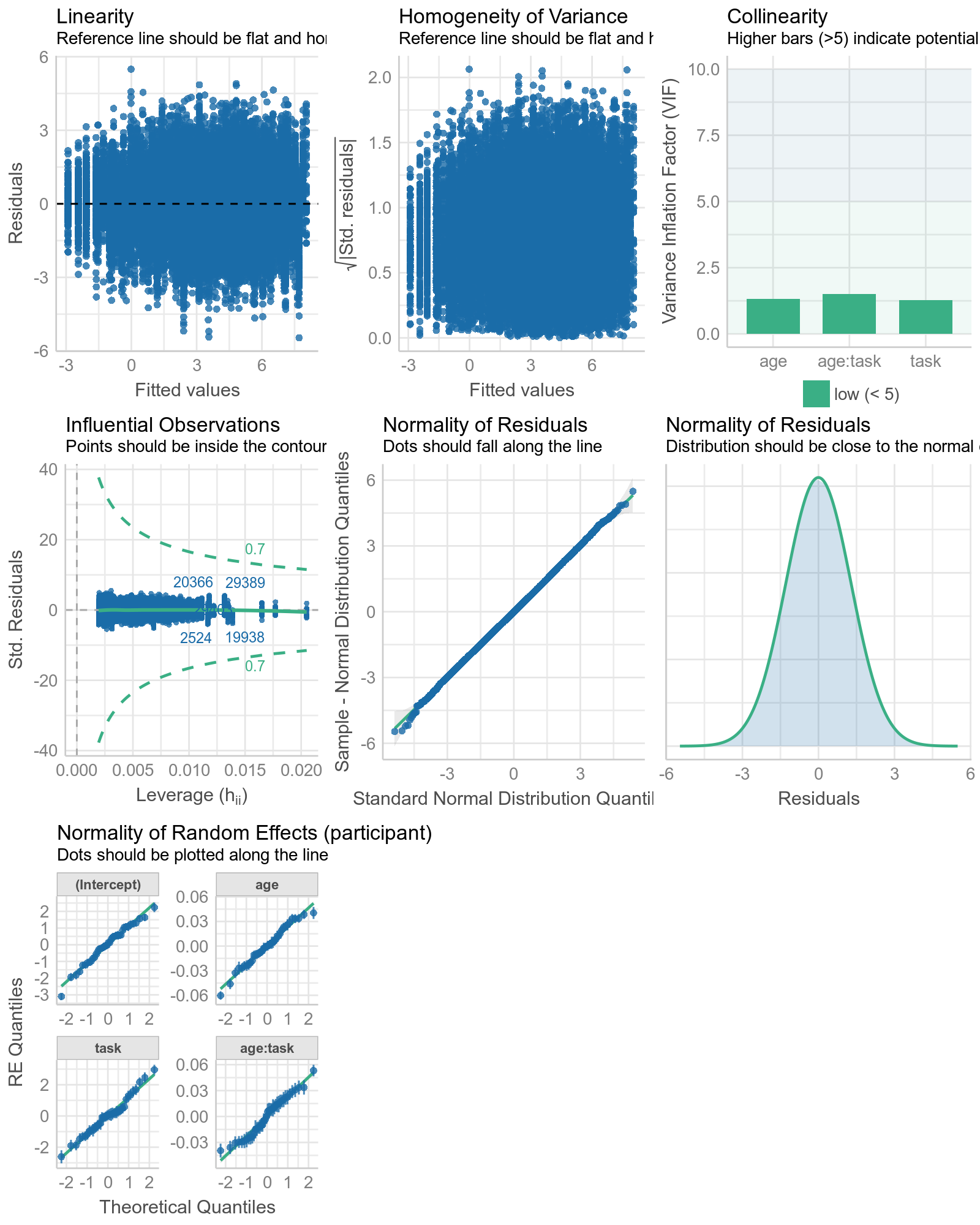
Other sources of randomness¶
A cross random factor mixed model E.g. if each trial had a different wm test/stimuli
random_mu_trial = rnorm(50, 0, 1)#A simple random effect where each trial has its own mean
varcovmat = matrix(c(1, .33, .33, .33, .33, 1, .33, .33, .33, .33,1,.33, .33, .33, .33,1), nrow=4)
data = data.frame()
for(i in 1:n_participants){
re = mvrnorm(n=1, mu=c(0, 0, 0, 0), Sigma=varcovmat)
mu_i = re[1] * sd_wm + mean_wm
b_age = re[2] *(sd_wm*2) - (sd_wm*4) #b_age_j
b_task = re[3] * sd_wm + -(sd_wm*2)#b_task_j
b_age_task = re[4] * (sd_wm/2) - (sd_wm*2)#b_agetask_j
age_tested = runif(runif(1, 5,15), 1, 99)
for (age in age_tested){
age = age/100
for (task in c(0,1)){
ntrials = 50
subdata = data.frame(trial=1:ntrials)
mu = mu_i+b_age*age+b_task*task+b_age_task*age*task
subdata$wm = rnorm(50, mu, sd=sd_wm)
subdata$wm = subdata$wm + rnorm(50, random_mu_trial, 1)#drawing 1 samples for the 50 trials with the 50 mu
subdata$participant = as.character(i)
subdata$age = age
subdata$task = task
subdata$trial = 1:50#trial n
data = rbind(data, subdata)
}
}
}
full_trial_fullFERE <- lmer(wm ~ task * age + (task * age|participant) + (1|trial), data = data,
control = lmerControl(optimizer = "optimx", calc.derivs = FALSE,
optCtrl = list(method = "nlminb", starttests = FALSE, kkt = FALSE)))
summary(full_trial_fullFERE)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: wm ~ task * age + (task * age | participant) + (1 | trial)
Data: data
Control:
lmerControl(optimizer = "optimx", calc.derivs = FALSE, optCtrl = list(method = "nlminb",
starttests = FALSE, kkt = FALSE))
REML criterion at convergence: 52240.6
Scaled residuals:
Min 1Q Median 3Q Max
-4.6949 -0.6614 0.0035 0.6593 3.9869
Random effects:
Groups Name Variance Std.Dev. Corr
trial (Intercept) 0.6099 0.7809
participant (Intercept) 1.0478 1.0236
task 1.0811 1.0397 0.37
age 5.0708 2.2518 0.62 0.55
task:age 0.2799 0.5290 0.13 0.24 0.52
Residual 1.9822 1.4079
Number of obs: 14700, groups: trial, 50; participant, 15
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 6.2142 0.2891 18.8944 21.495 9.67e-15 ***
task -1.9749 0.2738 14.1434 -7.212 4.23e-06 ***
age -4.0557 0.5856 14.0920 -6.926 6.78e-06 ***
task:age -1.5085 0.1661 14.2018 -9.080 2.72e-07 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) task age
task 0.309
age 0.549 0.545
task:age 0.142 0.095 0.381
plot_model(full_trial_fullFERE, type = "pred", terms = c("age","task"), show.data = TRUE)
options(repr.plot.width=10, repr.plot.height=10, repr.plot.res = 100)#hidden code for display size
plot_model(full_trial_fullFERE, type = "re")
options(repr.plot.width=8, repr.plot.height=5, repr.plot.res = 200)#hidden code for display size
[[1]] [[2]]
Final words for Module 2¶
Practical advices on LMM :
- Watch for the difference in scale between your predictors (e.g. we rescaled age factor between 0 and 1)
- Carefully think about the contrast of your predictor (centering predictor often helps some convergence problems)
- Be certain to have enough units in the RE (Gelman and Hill, 2007)
- By default, statistical packages such as lme4 assume a normal distribution of these RE
- Think about the relationship between your units (e.g. electrodes in an EEG)
If you want to learn more about simulation, mixed models and e.g. power calculation see DeBruine and Barr 2021
When using mixed models it is advocated to always keep the random structure maximal (Barr et al., 2013) but using real data we often end up in computational problems in the estimation. Other language (e.g. Julia) ? Bayesian estimation ?
Moving to module 3 after some practical exercices on the data you brought