Caution
- High statistical content taught by a non-statician
- Ask for guidance/consulting
- R code is kept simple -> sometimes unefficient (+ instructor = pythonista)
Organization of the course¶
- Module 1 : Linear models and necessary "reminders"
- Module 2 : Hierarchies in the data and mixed models
- Module 3 : Bayesian estimation and Generalized Linear Mixed Models
Why are we interested in linear models¶
When we can perform, t-tests, ANOVA, MANOVA, ANCOVA, $\chi^2$ ?
Because each of these statistical tests boil down to linear models
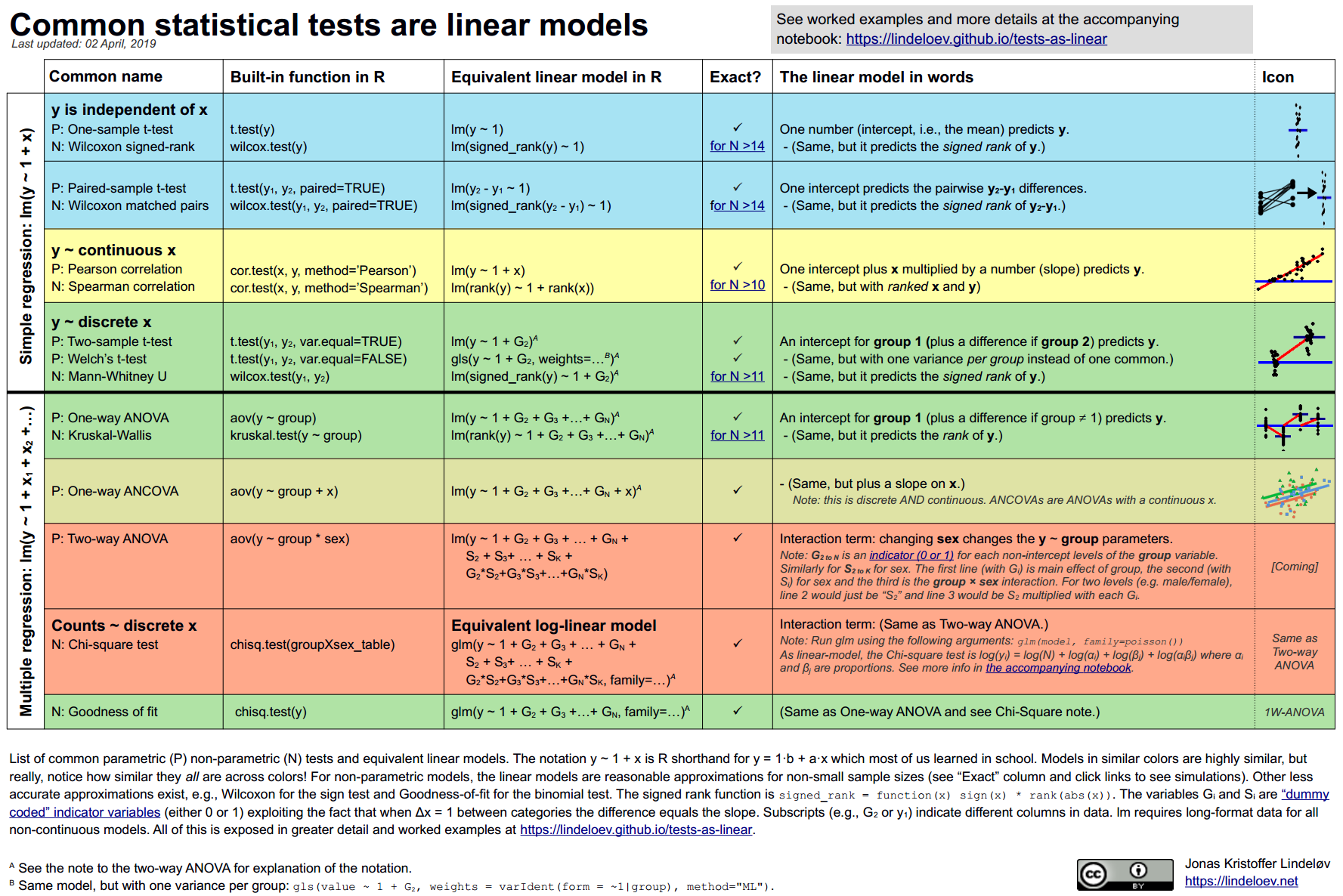
Linear Mixed Models are extensions of the linear models allowing to account for hierarchies in the data (e.g. participants, electrodes, labs, etc.).
First we are going to refresh your memories on linear models and build on that knowledge to talk about linear mixed models !
Organization of the first module¶
- Generalities on linear models
- Implementing a simple linear model in R + plotting the model and its assumptions
- Moving to LM with multiple predictiors
- Collinearity between predictors
- Contrast of the predictors
Generalities on linear models¶
A normal distribution is defined by a mean/location ($\mu$) and a standard deviation/scale ($\sigma$)
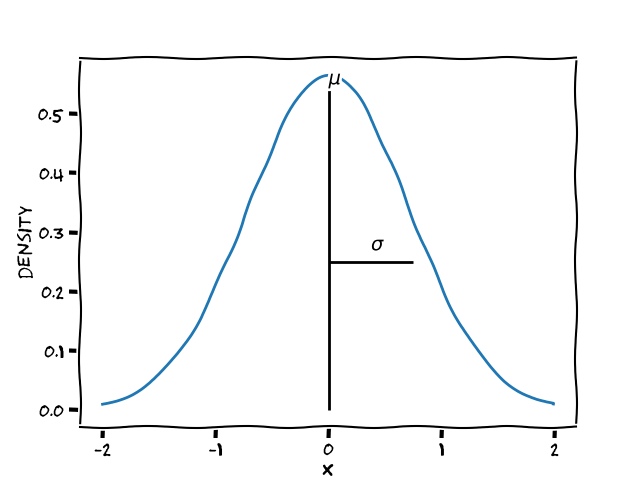
(Classical) Linear models are built on top of such distributions, e.g. for the normal distribution
The classical formula :
$y = \alpha + \beta IV + e$
Where :
$e \sim \mathcal{N}(0,\sigma)$
(Classical) Linear models are built on top of such distributions, e.g. for the normal distribution

Can be rewritten as :
$y \sim \mathcal{N}(\mu,\sigma)$
Where :
$\mu = \alpha + \beta IV$
$y \sim \mathcal{N}(\mu,\sigma) ~~~~~~\text{where}~~~ \mu = \alpha + \beta IV$
Provides an overview of some of the assumptions :
- Only $\mu$ is function of the IV -> homoscedasticity
- data points are independently and identically distributed i.i.d.
- The residuals are normally distributed
Two parameters for $\mu$ :
- $\alpha$ the intercept
- $\beta$ the slope
plot(0,0)
abline(a=1,b=0)
Let's draw some data points and see how regression line evolves:
https://observablehq.com/@tmcw/bring-your-own-doodles-linear-regression
Parameter estimation¶
- Ordinary least square (you know that one) :
- Maximum Likelihood Estimation (see excellent chapter/paper by Myung 2016) :
Note that we should go through linear algebra to have the complete picture of the math behind OLS and LM
Implementation in R¶
To implement linear models we use the base R function of lm().
Let's use an example :
set.seed(234)#I don't like randomness in my lectures
obs = 1000 #We sampled the WM of 100 persons
mean_wm = 6 #magical number 6
sd_wm = 1 #+/- 1
wm_v = rnorm(obs, mean_wm,sd_wm)#Random number generator
youngest = 1
oldest = 99
age_v = runif(obs, youngest,oldest)#Random number generator
data = data.frame(wm_v, age_v)
plot(data$age_v, data$wm_v, xlab= "Age",ylab="WM span")
But we want data with a linear relationship between DV (WM span) and the IV (age) :
b_age = -0.04 #aging of one unit (e.g. year) decreases the WM of 0.4 unit
data$wm_v = data$wm_v + b_age * data$age_v
plot(age_v, wm_v + (age_v*b_age), xlab="Age (year)", ylab="WM span")
#above is an operation on vector ;
# for each indiv i : wm_i = wm_v[i] + (age_v[i] * b_age)
Let's fit a lm¶
model <- lm(wm_v ~ age_v, data=data)
summary(model)
Call:
lm(formula = wm_v ~ age_v, data = data)
Residuals:
Min 1Q Median 3Q Max
-2.96585 -0.64057 0.04763 0.64576 3.14293
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 5.891183 0.062811 93.79 <2e-16 ***
age_v -0.038256 0.001091 -35.07 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.9842 on 998 degrees of freedom
Multiple R-squared: 0.552, Adjusted R-squared: 0.5516
F-statistic: 1230 on 1 and 998 DF, p-value: < 2.2e-16
Coefficients are not perfect , but we could increase the number of observations (e.g. 1000)
Now remember we should always plot the data and the models prediction

model$coeff#accessing coefficients
- (Intercept)
- 5.89118292468627
- age_v
- -0.0382562780830815
#plot(data$age_v, data$wm_v)
#abline(model$coeff[1], model$coeff[2], lwd=3, col="blue")
#We can even extend the prediction :
plot(data$age_v, data$wm_v,xlim = c(1,150))
abline(model$coeff[1], model$coeff[2], lwd=3, col="blue")
Next step we want to check the models assumption :
We could do this by hand
resid = data$wm_v - (model$coeff[1] + model$coeff[2] * data$age_v)
hist(resid)
But in R you have a package for a lot of things :
require("performance")#Useful automated assumption check package
check_model(model)
Loading required namespace: qqplotr For confidence bands, please install `qqplotr`.
Now what if we feed the wrong distribution ?¶
data_exp = data
data_exp$wm_v = exp(data_exp$wm_v)
exp_model <- lm(wm_v ~ age_v, data=data_exp)
summary(exp_model)
Call:
lm(formula = wm_v ~ age_v, data = data_exp)
Residuals:
Min 1Q Median 3Q Max
-329.31 -105.46 -25.38 51.80 2610.69
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 371.9083 14.3770 25.87 <2e-16 ***
age_v -4.5319 0.2497 -18.15 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 225.3 on 998 degrees of freedom
Multiple R-squared: 0.2481, Adjusted R-squared: 0.2474
F-statistic: 329.4 on 1 and 998 DF, p-value: < 2.2e-16
plot(data_exp$age_v, data_exp$wm_v)
abline(exp_model$coeff[1], exp_model$coeff[2], lwd=3, col="blue")
check_model(exp_model)
Loading required namespace: qqplotr For confidence bands, please install `qqplotr`.
Moving to linear models with multiple predictors¶
Let's add another predictor, e.g. with or without a simultaneous task to the working memory task
mean_double_task = 2
wm_v = rnorm(obs, mean_double_task,sd_wm)#Different mean
data2 = data.frame(wm_v, age_v)#age is the same
# Inducing an interaction :
b_age_double_task = -0.02 #aging of one unit (e.g. year) decreases the WM of 0.2 unit when double
data2$wm_v = data2$wm_v + b_age_double_task * data2$age_v
#Merging
data$condition = "single"
data2$condition = "double"
data_int = rbind(data, data2)
subset = data_int[data_int$condition == "single",]#lisibility/clarity purpose
plot(subset$age_v, subset$wm_v, xlab="Age (year)", ylab="WM span", col="red")
subset = data_int[data_int$condition == "double",]#lisibility/clarity purpose
points(subset$age_v, subset$wm_v, xlab="Age (year)", ylab="WM span", col="blue")
We can fit the model without interaction :
Now the formula is : $\mu = \alpha + \beta_{age} * Age + \beta_{task} * Task$
model_add <- lm(wm_v ~ age_v + condition, data=data_int)
Let's first plot our model using an automated R library
#install.packages("sjPlot")
library("sjPlot")
library(sjlabelled)
library(sjmisc)
library(ggplot2)
plot_model(model_add, type = "pred", terms = c("age_v", "condition"),
show.data = TRUE, ci.lvl = NA, line.size=3)
R's default behavior default coded single as = 1 and double as = 0 (but see later), let's print the output
summary(model_add)
Call:
lm(formula = wm_v ~ age_v + condition, data = data_int)
Residuals:
Min 1Q Median 3Q Max
-3.4833 -0.7029 0.0307 0.7009 3.2708
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.4564125 0.0520062 47.23 <2e-16 ***
age_v -0.0292721 0.0008094 -36.17 <2e-16 ***
conditionsingle 2.9854702 0.0461787 64.65 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.033 on 1997 degrees of freedom
Multiple R-squared: 0.7332, Adjusted R-squared: 0.7329
F-statistic: 2744 on 2 and 1997 DF, p-value: < 2.2e-16
We can fit the model with interaction
Now the formula is : $\mu = \alpha + \beta_{age} * Age + \beta_{task} * Task + \beta_{age \times Task} * Age * Task$
model_int <- lm(wm_v ~ age_v * condition, data=data_int)#We switched to "*"
#Or more explicitely :
model_int <- lm(wm_v ~ age_v + condition + age_v:condition, data=data_int)#same as above
plot_model(model_int, type = "pred", terms = c("age_v", "condition"),
show.data = TRUE, ci.lvl = NA, line.size=3)
Now analyze the summary table
summary(model_int)
Call:
lm(formula = wm_v ~ age_v + condition + age_v:condition, data = data_int)
Residuals:
Min 1Q Median 3Q Max
-3.2730 -0.6653 0.0310 0.6679 3.1429
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.007112 0.063852 31.43 <2e-16 ***
age_v -0.020288 0.001109 -18.29 <2e-16 ***
conditionsingle 3.884071 0.090301 43.01 <2e-16 ***
age_v:conditionsingle -0.017968 0.001568 -11.46 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1 on 1996 degrees of freedom
Multiple R-squared: 0.7496, Adjusted R-squared: 0.7493
F-statistic: 1992 on 3 and 1996 DF, p-value: < 2.2e-16
But the default R contrast is not coherent with our research question
Building on contrast coding of factors¶
Until now :
- $\alpha$ = predicted WM when age 0
- $\beta_{age}$ Effect of age on WM when in double condition
- $\beta_{condition}$ difference in intercept when going from double to single task
- $\beta_{age \times condition}$ change in effect of age on WM when single task
We rather would like :
- $\alpha$ = predicted WM when age 1
- $\beta_{age}$ Effect of age on WM when in single condition (reference) and difference between lowest and highest age
- $\beta_{condition}$ difference in intercept when going from single to double task
- $\beta_{age \times condition}$ change in effect of age/double task on WM when double task/age
We then recode the data
data_int$age_v_recoded = (data_int$age_v - 1)/100
data_int$condition_recoded = ifelse(data_int$condition == "single", 0, 1)
head(data_int)
| wm_v | age_v | condition | age_v_recoded | condition_recoded | |
|---|---|---|---|---|---|
| <dbl> | <dbl> | <chr> | <dbl> | <dbl> | |
| 1 | 6.3150246 | 8.643629 | single | 0.076436287 | 0 |
| 2 | 2.6518688 | 32.378705 | single | 0.313787047 | 0 |
| 3 | 0.6671325 | 95.841536 | single | 0.948415365 | 0 |
| 4 | 6.7755663 | 17.391671 | single | 0.163916705 | 0 |
| 5 | 5.9695408 | 37.239943 | single | 0.362399432 | 0 |
| 6 | 6.0742064 | 1.648316 | single | 0.006483155 | 0 |
tail(data_int)
| wm_v | age_v | condition | age_v_recoded | condition_recoded | |
|---|---|---|---|---|---|
| <dbl> | <dbl> | <chr> | <dbl> | <dbl> | |
| 1995 | 0.6745658 | 34.98079 | double | 0.3398079 | 1 |
| 1996 | 3.6278110 | 12.01254 | double | 0.1101254 | 1 |
| 1997 | 0.9268044 | 92.42589 | double | 0.9142589 | 1 |
| 1998 | 0.7432773 | 12.68647 | double | 0.1168647 | 1 |
| 1999 | -0.6180795 | 50.42658 | double | 0.4942658 | 1 |
| 2000 | 0.3038832 | 98.45748 | double | 0.9745748 | 1 |
model_int_recoded <- lm(wm_v ~ age_v_recoded * condition_recoded, data=data_int)
summary(model_int_recoded)
Call:
lm(formula = wm_v ~ age_v_recoded * condition_recoded, data = data_int)
Residuals:
Min 1Q Median 3Q Max
-3.2730 -0.6653 0.0310 0.6679 3.1429
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 5.85293 0.06289 93.06 <2e-16 ***
age_v_recoded -3.82563 0.11090 -34.49 <2e-16 ***
condition_recoded -3.86610 0.08894 -43.47 <2e-16 ***
age_v_recoded:condition_recoded 1.79683 0.15684 11.46 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1 on 1996 degrees of freedom
Multiple R-squared: 0.7496, Adjusted R-squared: 0.7493
F-statistic: 1992 on 3 and 1996 DF, p-value: < 2.2e-16
Now we have meaningful parameters, e.g. :
WM scores decreased with age ($\beta_{age} = -0.04, SE = 0.001, p < .05$) as does the presence of a double task ($\beta_{task} = -1.71, SE = 0.07, p < .05$). An interaction was present between age and task ($\beta_{age \times task} = 0.02, SE = 0.002, p < .05)$.
Think about your contrast¶
For binary predictors we can choose between :
- "sum" contrast (-1, 1)
- "treatment" contrast (0,1)
The linear model is the same but the interpretation of the intercept changes :
| parameter | Sum | Treatment |
|---|---|---|
| Intercept | mean between both conditions | mean when standard condition |
| Slope | Difference between conditions | Difference between conditions |
data_int$condition_treatment = ifelse(data_int$condition =="single",0,1)
data_int$condition_sum = ifelse(data_int$condition =="single",-1,1 )
summary(lm(wm_v ~ age_v * condition_treatment, data=data_int))$coefficients
summary(lm(wm_v ~ age_v * condition_sum, data=data_int))$coefficients
| Estimate | Std. Error | t value | Pr(>|t|) | |
|---|---|---|---|---|
| (Intercept) | 5.89118292 | 0.063852450 | 92.26244 | 0.000000e+00 |
| age_v | -0.03825628 | 0.001109038 | -34.49500 | 6.297549e-205 |
| condition_treatment | -3.88407058 | 0.090301001 | -43.01249 | 1.327709e-286 |
| age_v:condition_treatment | 0.01796830 | 0.001568417 | 11.45633 | 1.783488e-29 |
| Estimate | Std. Error | t value | Pr(>|t|) | |
|---|---|---|---|---|
| (Intercept) | 3.94914763 | 0.0451505007 | 87.46631 | 0.000000e+00 |
| age_v | -0.02927213 | 0.0007842086 | -37.32697 | 9.007915e-232 |
| condition_sum | -1.94203529 | 0.0451505007 | -43.01249 | 1.327709e-286 |
| age_v:condition_sum | 0.00898415 | 0.0007842086 | 11.45633 | 1.783488e-29 |
Now imagine we have three modalities :
- single task
- double task same modality (verbal - verbal)
- double task different modalities (verbal - non-verbal)
mean_double_task_same = 5
mean_double_task_diff = 2
wm_v = rnorm(obs, mean_double_task_same,sd_wm)#Different mean
data_same = data.frame(wm_v, age_v)
wm_v = rnorm(obs, mean_double_task_diff,sd_wm)#Different mean
data_diff = data.frame(wm_v, age_v)
#Merging
data$condition = "a-single"
data_same$condition = "d-same"
data_diff$condition = "d-diff"
data_threemod = rbind(data, data_same, data_diff)
data_threemod$condition = as.factor(data_threemod$condition)#We are working with the factor class
print(levels(data_threemod$condition))
[1] "a-single" "d-diff" "d-same"
Sum contrasting your n modalities
data_threemod$condition_sum = data_threemod$condition
contrasts(data_threemod$condition_sum) = contr.sum(3)#Defining the contrast matrix
print(contrasts(data_threemod$condition_sum)) #printing it
sum_cont = lm(wm_v ~ age_v * condition_sum, data=data_threemod)
summary(sum_cont)$coefficients
[,1] [,2] a-single 1 0 d-diff 0 1 d-same -1 -1
| Estimate | Std. Error | t value | Pr(>|t|) | |
|---|---|---|---|---|
| (Intercept) | 4.31972552 | 0.0368106263 | 117.34996 | 0.000000e+00 |
| age_v | -0.01283485 | 0.0006393552 | -20.07468 | 3.356279e-84 |
| condition_sum1 | 1.57145741 | 0.0520580870 | 30.18661 | 5.407327e-175 |
| condition_sum2 | 0.72329773 | 0.0520580870 | 13.89405 | 1.403789e-42 |
| age_v:condition_sum1 | -0.02542142 | 0.0009041848 | -28.11529 | 1.508955e-154 |
| age_v:condition_sum2 | 0.01225058 | 0.0009041848 | 13.54875 | 1.239886e-40 |
Treatment contrasting your n modalities
data_threemod$condition_treatment = data_threemod$condition
contrasts(data_threemod$condition_treatment) = contr.treatment(3)
print(contrasts(data_threemod$condition_treatment))
treatment_cont = lm(wm_v ~ age_v * condition_treatment, data=data_threemod)
summary(treatment_cont)$coefficients
2 3 a-single 0 0 d-diff 1 0 d-same 0 1
| Estimate | Std. Error | t value | Pr(>|t|) | |
|---|---|---|---|---|
| (Intercept) | 5.89118292 | 0.063363297 | 92.97469 | 0.000000e+00 |
| age_v | -0.03825628 | 0.001100542 | -34.76129 | 1.036410e-222 |
| condition_treatment2 | -0.96246299 | 0.089609234 | -10.74067 | 1.974392e-26 |
| condition_treatment3 | -3.92146477 | 0.089609234 | -43.76184 | 9.881313e-324 |
| age_v:condition_treatment2 | 0.04044263 | 0.001556402 | 25.98469 | 2.067890e-134 |
| age_v:condition_treatment3 | 0.03937246 | 0.001556402 | 25.29710 | 4.009068e-128 |
But we can define custom contrasts
data_threemod$condition_sum = data_threemod$condition
contrasts(data_threemod$condition_sum,how.many=2) = matrix(c(-1,0,1,-1,1,0),ncol=2)
print(contrasts(data_threemod$condition_sum))
sum_cont = lm(wm_v ~ age_v * condition_sum, data=data_threemod)
summary(sum_cont)$coefficients
[,1] [,2] a-single -1 -1 d-diff 0 1 d-same 1 0
| Estimate | Std. Error | t value | Pr(>|t|) | |
|---|---|---|---|---|
| (Intercept) | 4.26320701 | 0.0365828164 | 116.53578 | 0.000000e+00 |
| age_v | -0.01165125 | 0.0006353985 | -18.33692 | 2.903960e-71 |
| condition_sum1 | -2.29348885 | 0.0517359152 | -44.33069 | 0.000000e+00 |
| condition_sum2 | 0.66551293 | 0.0517359152 | 12.86365 | 6.719274e-37 |
| age_v:condition_sum1 | 0.01276743 | 0.0008985891 | 14.20831 | 2.183827e-44 |
| age_v:condition_sum2 | 0.01383760 | 0.0008985891 | 15.39925 | 1.489765e-51 |
We could build more complex contrast coding schemes (see the paper by Schad, Vasishth, Hohenstein, Kliegl 2020 )
Collinearity¶
Multiple predictors yields the question of collinearity, i.e. when another factor does not add a lot of information compared to using only the first factor.
Example let's simulate data where we predict reaction time (RTs) using IQ and memory scores
library('MASS')
varcovmat = matrix(c(1, .97, .97, 1), nrow=2)
data = mvrnorm(n=100, mu=c(0, 0), Sigma=varcovmat, empirical=TRUE)
iq = data[, 1]*15 +100#Just rescaling for "natural" scale
mem = data[, 2]*15 +100 #same
plot(iq,mem)
rt = rnorm(100, 500,50) + -1*iq #Only relationship simulated is with IQ
plot(iq, rt)
model_rt = lm(rt ~ iq + mem)#Additive, we did not include an interaction
summary(model_rt)
Call:
lm(formula = rt ~ iq + mem)
Residuals:
Min 1Q Median 3Q Max
-107.392 -31.466 -1.463 24.007 94.377
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 483.2304 29.1289 16.589 <2e-16 ***
iq -0.7652 1.1764 -0.651 0.517
mem -0.0725 1.1764 -0.062 0.951
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 42.68 on 97 degrees of freedom
Multiple R-squared: 0.08092, Adjusted R-squared: 0.06197
F-statistic: 4.27 on 2 and 97 DF, p-value: 0.0167
check_collinearity(model_rt)
| Term | VIF | SE_factor | |
|---|---|---|---|
| <chr> | <dbl> | <dbl> | |
| 1 | iq | 16.92047 | 4.11345 |
| 2 | mem | 16.92047 | 4.11345 |
model_rt = lm(rt ~ iq)
summary(model_rt)
Call:
lm(formula = rt ~ iq)
Residuals:
Min 1Q Median 3Q Max
-107.806 -31.207 -1.267 23.974 93.890
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 483.0129 28.7670 16.791 < 2e-16 ***
iq -0.8356 0.2845 -2.937 0.00413 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 42.46 on 98 degrees of freedom
Multiple R-squared: 0.08089, Adjusted R-squared: 0.07151
F-statistic: 8.624 on 1 and 98 DF, p-value: 0.004134
But collinearity is not a pathology of the model (e.g. IQ and memory indices are generally correlated)
Statistical inference on LM¶
- Use stepwise regressions : how to do it in R
Statistical inference on LM¶
Limitations of the classical LM¶
Problems is that in psychology/neuroscience :
- Within design (e.g. same participant performing single and double task) -> Non iid data
- Repeated measurment (e.g. we sample several scores for precision)-> Non iid data
- Non-normal data (e.g. Reaction times)
The last two points could be alleviated by taking a central tendency index (e.g. mean) but we would loose information.
Using Linear Mixed Models¶
- Directly allows to account for within design -> preserve the higher precision of within design and accounting for the non i.i.d nature induced by within design
- Account for repeated measurment -> no pre-averaging
- Imbalance in repeated measure (e.g. one participant with fewer trials) is compensated by LMM
- Provides estimates in terms of effect size rather than obscure data-abstract statistical values (t, F)
But... there is no free lunch
